Ohio State, University of Washington scientists pave way for protein nanomachines
While the Ohio State Buckeyes and Washington Huskies prepared to butt heads in the Rose Bowl on Jan. 1, researchers at the two universities collaborated to create proteins that zip together in much the same way DNA molecules zip up to form a double helix.
The first-of-its-kind scientific breakthrough could enable the development of protein nanomachines that could help diagnose and treat disease, allow for more exact engineering of cells and perform a wide variety of other tasks.
“This technique makes it possible for you to design proteins so they come together exactly how you want them to,” said Zibo Chen, a graduate student at the University of Washington and lead author of the paper, which was published Dec. 19 in the journal Nature.
The research heavily relied on the technical expertise of Ohio State’s Resource for Native Mass Spectrometry Guided Structural Biology, an NIH-funded center led by Vicki Wysocki, professor of chemistry and biochemistry, Ohio Eminent Scholar and director of the Campus Chemical Instrument Center.
“Our contribution was the analysis of protein designs to confirm they were pairing correctly,” said Florian Busch, an Ohio State biochemist at the Resource for Native Mass Spectrometry and an author on the paper. “We also co-evolved with the ongoing project and came up with methods to analyze the designs faster and in mixtures that allowed proteins to find their correct partners even in the presence of other similar proteins.”
Typically, researchers interested in designing biomolecular nanomachines use DNA as the primary component, since complementary DNA strands come together in a highly predictable manner determined by base-pairing. The team developed protein designs that produce complementary proteins that precisely pair with each other by a similar approach, Busch explained.
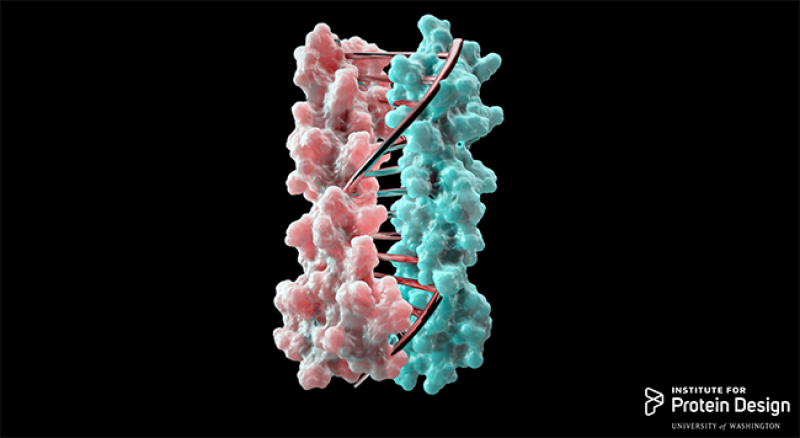
“It’s like you have two pieces of a zipper … You can find the pairs in the zipper, zip them together, combine more pieces into a large complex and then finally assemble some functional proteins,” said Mengxuan Jia, an Ohio State graduate student in the Department of Chemistry and Biochemistry and collaborator on the study.
The University of Washington side of the team designed the new proteins computationally, using hydrogen-bond networks to ensure that each protein pair had a unique complementary sequence that wouldn’t cross-react with proteins from other pairs.
“Engineering cells to do new tasks is the future of medicine and biotechnology, whether that’s engineering bacteria to make energy or clean up toxic waste or creating immune cells that attack cancers,” said Scott Boyken, a co-author of the paper and postdoctoral researcher at UW Medicine’s Institute for Protein Design. “We have opened a major door to protein nanomaterial design.”
Overall, the new findings pave the way for transforming biomedical technology and biomolecular engineering.
“Almost all biological processes depend on protein-protein interaction,” Busch said. “Our next steps will be working with bigger, more dynamic protein assemblies and on understanding how more complicated complexes assemble and disassemble.”
.dailypost {background-color:#000; padding:30px;color:#fff;font-family:"capita";font-size: 1.25em;font-weight: 400;} .clicktotweet {float: right; text-align:right;}
Scientists program proteins to pair exactly. #ASCDaily
The ability to test and confirm the new designs and their interactions is a testament to the unique power of native mass spectrometry, which can provide highly detailed information about protein complexes while keeping them intact and as close to their native state as possible.
“This is a prime example of how collaboration to address questions that have both biomedical significance and substantial technical challenges drives innovation in our newly established Resource for Native Mass Spectrometry Guided Structural Biology,” Wysocki said. The Resource is currently working on around 12 such biomedical projects with investigators across the nation and globe.
This story includes information from a University of Washington press release. Zachary L. VanAernum and Aniruddha Sahasrabuddhe, current and former graduate students, respectively, at Ohio State’s Department of Chemistry and Biochemistry, were also involved in this study.
